Publications
Explore our latest scientific articles, research papers, and contributions to advance knowledge in the field.
BOOKS
MACHINE LEARNING
The Goldilocks paradigm: comparing classical machine learning, large language models, and few-shot learning for drug discovery applications

Assessment of Substrate-Dependent Ligand Interactions at the Organic Cation Transporter OCT2 Using Six Model Substrates

Computational drug repositioning identifies niclosamide and tribromsalan as inhibitors of Mycobacterium tuberculosis and Mycobacterium abscessus

Machine learning-aided search for ligands of P2Y6 and other P2Y receptors

Generative Artificial Intelligence-Assisted Protein Design Must Consider Repurposing Potential

High-Throughput Phenotypic Screening and Machine Learning Methods Enabled the Selection of Broad-Spectrum Low-Toxicity Antitrypanosomatidic Agents

There’s a ‘ChatGPT’ for biology. What could go wrong?

Comparing LD50/LC50 Machine Learning Models for Multiple Species

Validation of Acetylcholinesterase Inhibition Machine Learning Models for Multiple Species

Stakeholder perspectives on the Biological Weapons Convention

A teachable moment for dual-use.

Confirmation of high-throughput screening data and novel mechanistic insights into FXR-xenobiotic interactions by orthogonal assays

Al-novation: Finding New Uses for Artificial Intelligence Across Industries

AI in drug discovery: A wake-up call

Machine Learning Models Identify New Inhibitors for Human OATP1B1.

Combining DELs and machine learning for toxicology prediction.

Machine Learning for Discovery of New ADORA Modulators.

Rickettsia Aglow: A Fluorescence Assay and Machine Learning Model to Identify Inhibitors of Intracellular Infection.

Integrating Generative Molecular Design, Automated Analog Designer, and Synthetic Viability Prediction

The Commoditization of AI for Molecule Design.

Dual use of artificial-intelligence-powered drug discovery

Mycobacterium abscessus drug discovery using machine learning

Machine Learning Models for Mycobacterium tuberculosis In VitroActivity: Prediction and Target Visualization

UV-adVISor: Attention-Based Recurrent Neural Networks to Predict UV-Vis Spectra

Remdesivir and EIDD-1931 Interact with Human Equilibrative Nucleoside Transporters 1 and 2: Implications for Reaching SARS-CoV-2 Viral Sanctuary Sites

Machine Learning Models Identify Inhibitors of SARS-CoV-2

Bayesian Modeling and Intrabacterial Drug Metabolism Applied to Drug-Resistant Staphylococcus aureus

Development of Machine Learning Models and the Discovery of a New Antiviral Compound against Yellow Fever Virus

Recent advances in drug repurposing using machine learning.

Cationic Compounds with SARS-CoV-2 Antiviral Activity and their Interaction with OCT/MATE Secretory Transporters

Comparing the Pfizer Central Nervous System Multiparameter Optimization Calculator and a BBB Machine Learning Model

Quantum Machine Learning Algorithms for Drug Discovery Applications

Multiple Computational Approaches for Predicting Drug Interactions with Human Equilibrative Nucleoside Transporter 1

CATMoS: Collaborative Acute Toxicity Modeling Suite

Discovery of 5-Nitro-6-thiocyanatopyrimidines as Inhibitors of Cryptococcus neoformans and Cryptococcus gattii

A Machine Learning Strategy for Drug Discovery Identifies Anti-Schistosomal Small Molecules

Bioactivity Comparison across Multiple Machine Learning Algorithms Using over 5000 Datasets for Drug Discovery.

Comparing Machine Learning Models for Aromatase (P450 19A1).

Predicting Drug Interactions with Human Equilibrative Nucleoside Transporters 1 and 2 Using Functional Knockout Cell Lines and Bayesian Modeling.

Computational Approaches to Identify Molecules Binding to Mycobacterium Tuberculosis KasA

Comparison of Machine Learning Models for the Androgen Receptor

Machine Learning for Discovery of GSK3β Inhibitors

Evaluation of Assay Central® Machine Learning Models for Rat Acute Oral Toxicity Prediction

Machine Learning Models for Estrogen Receptor Bioactivity and Endocrine Disruption Prediction

Synergistic drug combinations and machine learning for drug repurposing in chordoma.

Machine Learning Platform to Discover Novel Growth Inhibitors of Neisseria gonorrhoeae.

Comparing Machine Learning Algorithms for Predicting Drug-Induced Liver Injury (DILI).

Pruned Machine Learning Models to Predict Aqueous Solubility
Cheminformatics Analysis and Modeling with MacrolactoneDB

Multiple Machine Learning Comparisons of HIV Cell-based and Reverse Transcriptase Data Sets.

Ebola Virus Bayesian Machine Learning Models Enable New in Vitro Leads.

Halogen Substitution Influences Ketamine Metabolism by Cytochrome P450 2B6: In Vitro and Computational Approaches.

Opportunities and Challenges Using Artificial Intelligence in ADME/Tox

Exploiting Machine Learning for End-to-End Drug Discovery and Development

High-Throughput Screening and Bayesian Machine Learning for Copper-Dependent Inhibitors of Staphylococcus Aureus
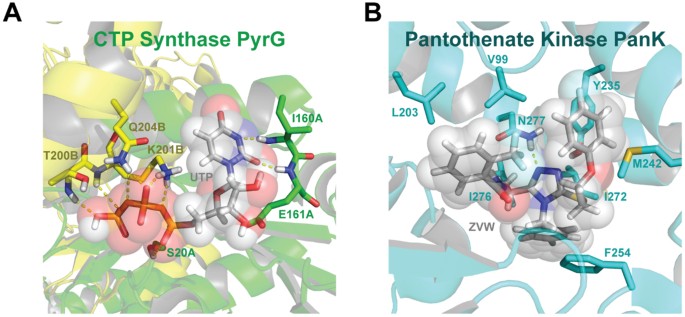
A multitarget approach to drug discovery inhibiting Mycobacterium tuberculosis PyrG and PanK.

Comparing and Validating Machine Learning Models for Mycobacterium tuberculosis Drug Discovery

The EU approved antimalarial pyronaridine shows antitubercular activity and synergy with rifampicin, targeting RNA polymerase.

High Throughput and Computational Repurposing for Neglected Diseases.

Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction

Comparison of Deep Learning With Multiple Machine Learning Methods and Metrics Using Diverse Drug Discovery Data Sets
ANTIVIRALS

1-Sulfonyl-3-amino-1H-1,2,4-triazoles as Yellow Fever Virus Inhibitors: Synthesis and Structure-Activity Relationship

Synthesis and Evaluation of 9-Aminoacridines with SARS-CoV-2 Antiviral Activity

Antiviral Evaluation of Dispirotripiperazines against Hepatitis B Virus

Learning from COVID-19: How drug hunters can prepare for the next pandemic

Discovery of PLpro and Mpro Inhibitors for SARS-CoV-2

Efficacy of an isoxazole-3-carboxamide analog of pleconaril in mouse models of Enterovirus-D68 and Coxsackie B5

N-Phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine as a New Class of HIV-1 Non-nucleoside Reverse Transcriptase Inhibitor.

Transporter Inhibition Profile for the Antivirals Tilorone, Quinacrine and Pyronaridine

The protein disulfide isomerase inhibitor 3-methyltoxoflavin inhibits Chikungunya virus.

Discovery of New Zika Protease and Polymerase Inhibitors through the Open Science Collaboration Project OpenZika

Vandetanib Blocks the Cytokine Storm in SARS-CoV-2-Infected Mice

Computational and Experimental Approaches Identify Beta-Blockers as Potential SARS-CoV-2 Spike Inhibitors

Pyronaridine Protects against SARS-CoV-2 Infection in Mouse

The need for speed and efficiency: A brief review of small molecule antivirals for COVID-19

Chalcones from Angelica keiskei (ashitaba) inhibit key Zika virus replication protein

Defending Antiviral Cationic Amphiphilic Drugs That May Cause Drug-Induced Phospholipidosis

Repurposing the Ebola and Marburg Virus Inhibitors Tilorone, Quinacrine and Pyronaridine: In Vitro Activity Against SARS-CoV-2 and Potential Mechanisms

The Antiviral Drug Tilorone Is A Potent and Selective Inhibitor of Acetylcholinesterase

Flavonoids from Pterogyne nitens as Zika virus NS2B-NS3 protease inhibitors

The Past, Present and Future of RNA Respiratory Viruses: Influenza and Coronaviruses

déjà vu: Stimulating Open Drug Discovery for SARS CoV 2

Repurposing Quaternary Ammonium Compounds as Potential Treatments for COVID-19

Pyronaridine Tetraphosphate Efficacy Against Ebola Virus Infection in Guinea Pig.

Toward the Target: Tilorone, Quinacrine, and Pyronaridine Bind to Ebola Virus Glycoprotein

Repurposing Pyramax®, quinacrine and tilorone as treatments for Ebola virus disease

Dispirotripiperazine-core compounds, their biological activity with a focus on broad antiviral property, and perspectives in drug design

Repurposing Quinacrine against Ebola Virus Infection In Vivo

Repurposing the antimalarial pyronaridine tetraphosphate to protect against Ebola virus infection.

The Natural Product Eugenol Is an Inhibitor of the Ebola Virus In Vitro
RARE OR NEGLECTED DISEASES

Identification of New Modulators and Inhibitors of Palmitoyl-Protein Thioesterase 1 for CLN1 Batten Disease and Cancer

Developing treatments for rare diseases on a shoestring

Multiple approaches to repurposing drugs for neuroblastoma

Cross-species efficacy of enzyme replacement therapy for CLN1 disease in mice and sheep

Advancing the Research and Development of Enzyme Replacement Therapies for Lysosomal Storage Diseases

Knowledge-based approaches to drug discovery for rare diseases.

Using Bibliometric Analysis and Machine Learning to Identify Compounds Binding to Sialidase-1

Repurposing the Dihydropyridine Calcium Channel Inhibitor Nicardipine as a Nav1.8 Inhibitor In Vivo for Pitt Hopkins Syndrome

Repurposing Approved Drugs as Inhibitors of Kv7.1 and Nav1.8 to Treat Pitt Hopkins Syndrome

Doing it All - How Families are Reshaping Rare Disease Research

Industrializing rare disease therapy discovery and development

Incentives for starting small companies focused on rare and neglected diseases

Collaboration for rare disease drug discovery research

Recommendations to enable drug development for the inherited neuropathies: Charco-Marie-Tooth and Giant Axonal Neuropathy

The multifaceted roles of rare disease parent / patient advocates in drug discovery

A generalizable pre-clinical research approach for orphan disease therapy
TOXICOLOGY



